Issues_ebpmf_rankk
zihao12
2019-10-06
Last updated: 2019-10-06
Checks: 7 0
Knit directory: ebpmf_demo/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190923) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/ebpmf_demo.Rmd
Untracked: analysis/ebpmf_rank1_demo2.Rmd
Untracked: analysis/softmax_experiments.ipynb
Untracked: data/ebpmf_issue.Rds
Untracked: data/ebpmf_issue2.Rds
Untracked: docs/figure/Experiment_ebpmf_rankk.Rmd/
Untracked: docs/figure/test.Rmd/
Unstaged changes:
Modified: analysis/ebpmf_rank1_demo.Rmd
Modified: analysis/ebpmf_rankk_demo.Rmd
Modified: analysis/softmax_experiments.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 5d1ad79 | zihao12 | 2019-10-06 | fix issue 2 |
| html | 42a291a | zihao12 | 2019-10-06 | Build site. |
| Rmd | bd74876 | zihao12 | 2019-10-06 | update issue |
| html | 66b65bd | zihao12 | 2019-10-06 | Build site. |
| Rmd | e733628 | zihao12 | 2019-10-06 | update issue |
| html | 73de8a3 | zihao12 | 2019-10-06 | Build site. |
| Rmd | 4eba31d | zihao12 | 2019-10-06 | update issue |
| html | 46742b0 | zihao12 | 2019-10-06 | Build site. |
| Rmd | 0336875 | zihao12 | 2019-10-06 | issues encountered in ebpmf |
Experiment setup
I put my code for https://zihao12.github.io/ebpmf_demo/ebpmf_rankk_demo2.html into a pacakge: https://github.com/zihao12/ebpmf
It is the same as in https://zihao12.github.io/ebpmf_demo/ebpmf_rankk_demo2.html except that I record the ELBO function (from
ebpmf::ebpmf_exponential_mixture_experimentfunction) and find a few issues.
Issues:
ELBO does not strictly increase. I think, if we can really solve each
ebpmproblem optimally within that prior family \(\mathcal{G}\), we should mamke ELBO nondecreasing (which is why coordinate ascend works?). But since in practice, we only optimize it over a subset of \(\mathcal{G}\) (based on our chosen grid), we may not solve each subproblem well.[solved] Numerical issue when computing
L: at iteration 107, I getError in verify.likelihood.matrix(L) : Input argument "L" should be a numeric matrix with >= 2 columns, >= 1 rows, all its entries should be non-negative, finite and not NA, and some entries should be positive. It happens inebpmwhen computing \(l_{ik} = log \ NB(x_i; a_k, \frac{b_k}{b_k + s})\). Only the first column of l (corresponding to the smallest chosen mean (biggest rate)) are all -inf. Why it happens? This is how we choose the gridxprime = x xprime[x == 0] = xprime[x == 0] + 1 mu_grid_min = 0.05*min(xprime/s) mu_grid_max = 2*max(x/s)In this case, the smallest nonzero \(x\) is very small, so
mu_grid_minbecomes very small (\(10^{-18}\)). As a result, the loglikelihood is-Inffor nonzero \(x\) (which is fine, asexp(-Inf) = 0), andNANfor zeros. The latter is strange as my code does not handlebig - smallnumber very well, and R producesNANfor0*-Inf:x = 0; prob = 1 - 1e-20; x*log(1-prob) ## [1] NaNI use this for my implementation of the “continous” negative binomial loglikelihood. I solve it (probably not the best way) by setting that term to 0 “manually”.
I have solved issue 2 and reproduce issue 1 here.
## Experiments
library(NNLM)
library(gtools)
sim_mgamma <- function(dist){
pi = dist$pi
a = dist$a
b = dist$b
idx = which(rmultinom(1,1,pi) == 1)
return(rgamma(1, shape = a[idx], rate = b[idx]))
}
## simulate a poisson mean problem
## to do:
simulate_pm <- function(n, p, dl, df, K,scale_b = 10, seed = 123){
set.seed(seed)
## simulate L
a = replicate(dl,1)
b = 10*runif(dl)
pi <- rdirichlet(1,rep(1/dl, dl))
gl = list(pi = pi, a = a, b= b)
L = matrix(replicate(n*K, sim_mgamma(gl)), ncol = K)
## simulate F
a = replicate(df,1)
b = 10*runif(df)
pi <- rdirichlet(1,rep(1/df, df))
gf = list(pi = pi, a = a, b= b)
F = matrix(replicate(p*K, sim_mgamma(gf)), ncol = K)
## simulate X
lam = L %*% t(F)
X = matrix(rpois(n*p, lam), nrow = n)
Y = matrix(rpois(n*p, lam), nrow = n)
## prepare output
g = list(gl = gl, gf = gf)
out = list(X = X, Y = Y, L = L, F = F, g = g)
return(out)
}Simulate data
### simulate data
n = 100
p = 200
K = 2
dl = 10
df = 10
scale_b = 5
sim = simulate_pm(n, p, dl, df, K, scale_b = scale_b)Run ebpmf and plot ELBO.
## ebpmf
out = ebpmf::ebpmf_exponential_mixture_experiment(sim$X, K, maxiter.out = 150, verbose = F)[1] "summary of runtime:"
[1] "init : 0.029000"
[1] "Ez per time: 0.001277"
[1] "rank1 per time: 0.025443"elbo_ebpmf= out$ELBO
out_ebpmf = out$qg
distance_max = max(elbo_ebpmf) - elbo_ebpmf
plot(1:length(distance_max), distance_max, xlab = "niter", ylab = "distance to max (ELBO)", type = "l", col = "blue")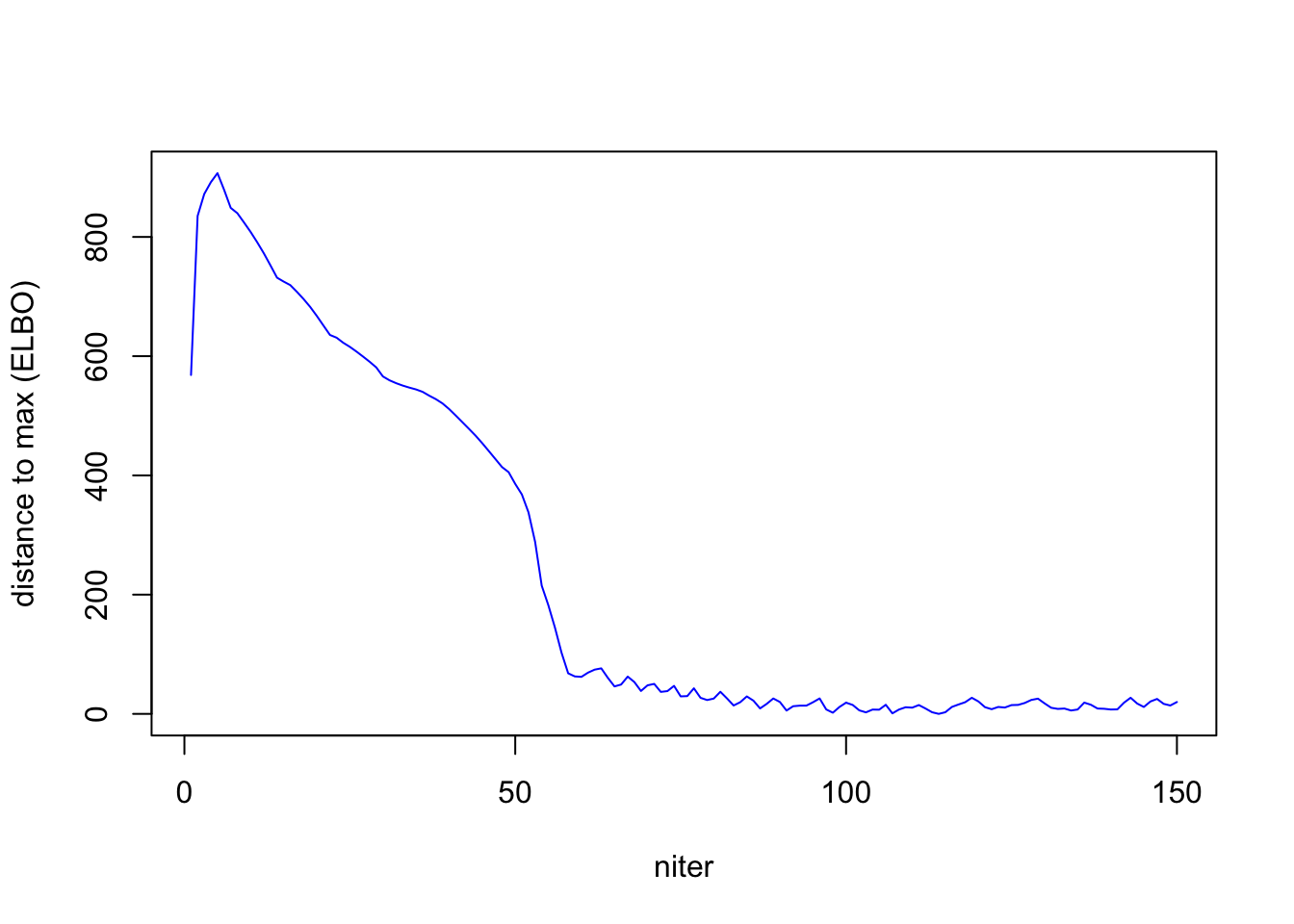
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] gtools_3.8.1 NNLM_0.4.2
loaded via a namespace (and not attached):
[1] workflowr_1.4.0 Rcpp_1.0.2 digest_0.6.21 rprojroot_1.3-2
[5] ebpmf_0.1.0 backports_1.1.5 git2r_0.25.2 magrittr_1.5
[9] evaluate_0.14 ebpm_0.0.0.9000 stringi_1.4.3 fs_1.3.1
[13] whisker_0.3-2 rmarkdown_1.13 tools_3.5.1 stringr_1.4.0
[17] glue_1.3.1 mixsqp_0.1-120 xfun_0.8 yaml_2.2.0
[21] compiler_3.5.1 htmltools_0.3.6 knitr_1.25