debug_ebpmf_point_gamma
zihao12
2019-10-22
Last updated: 2019-10-22
Checks: 7 0
Knit directory: ebpmf_demo/
This reproducible R Markdown analysis was created with workflowr (version 1.4.0). The Checks tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190923) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great job! Using relative paths to the files within your workflowr project makes it easier to run your code on other machines.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .Rhistory
Ignored: .Rproj.user/
Untracked files:
Untracked: analysis/.ipynb_checkpoints/
Untracked: analysis/ebpmf_demo.Rmd
Untracked: analysis/ebpmf_rank1_demo2.Rmd
Untracked: analysis/softmax_experiments.ipynb
Untracked: data/trash/
Untracked: docs/figure/Experiment_ebpmf_rankk.Rmd/
Untracked: docs/figure/test.Rmd/
Untracked: verbose_log_1571583163.21966.txt
Untracked: verbose_log_1571583324.71036.txt
Untracked: verbose_log_1571583741.94199.txt
Untracked: verbose_log_1571588102.40356.txt
Unstaged changes:
Modified: analysis/ebpmf_rank1_demo.Rmd
Modified: analysis/ebpmf_rankk_demo.Rmd
Modified: analysis/softmax_experiments.Rmd
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 7814be6 | zihao12 | 2019-10-22 | debug point gamma |
I managed to find the bug in ebpmf_point_gamma.Now ELBO increases monotonically.
Also noteworthy is the differences in RMSE compared with https://zihao12.github.io/ebpmf_demo/debug_ebpmf_exponential_mixture.html . Note the data generating process is almost the same. I would further compare the two ebpmf methods.
rm(list = ls())
devtools::load_all("../ebpmf")Warning: 1 components of `...` were not used.
We detected these problematic arguments:
* `action`
Did you misspecify an argument?Loading ebpmfWarning: package 'testthat' was built under R version 3.5.2set.seed(123)
library(NNLM)
library(ebpmf)
simulate_data <- function(n, p, K, params, seed = 1234){
set.seed(seed)
L = matrix(rgamma(n = n*K, shape = params$al, rate = params$bl), ncol = K)
F = matrix(rgamma(n = p*K, shape = params$af, rate = params$bf), ncol = K)
Lam = L %*% t(F)
X = matrix(rpois(n*p, Lam), nrow = n)
Y = matrix(rpois(n*p, Lam), nrow = n)
return(list(params = params,Lam = Lam,X = X, Y = Y))
}
n = 100
p = 200
K = 3
params = list(al = 100, bl = 109, af = 100, bf = 100, a = 1)
sim = simulate_data(n, p, K, params, seed = 1234)
maxiter = 100rank 1
mle
## MLE
mle_rank1 = NNLM::nnmf(A = sim$X, k = 1, method = "lee", loss = "mkl", max.iter = 1)
lam_mle_rank1 = mle_rank1$W %*% mle_rank1$H
ll_train_mle_rank1 = sum(dpois(sim$X, lam_mle_rank1, log = T))
ll_val_mle_rank1 = sum(dpois(sim$Y, lam_mle_rank1, log = T))
rmse_mle_rank1 = sqrt(mean((lam_mle_rank1 - sim$Lam)^2))
data.frame(ll_train = ll_train_mle_rank1, ll_val = ll_val_mle_rank1, rmse = rmse_mle_rank1) ll_train ll_val rmse
1 -37606.75 -37849.52 0.1978021ebpmf
## rank 1 case
out_rank1 = ebpmf::ebpmf_rank1_point_gamma_helper(X = sim$X, maxiter = 10, verbose = T)[1] "iter elbo kl_l kl_f sum_El sum_Ef"
[1] " 1 1525.9978611673 45.3663647810 68.4682771405 55800.5672468702 1.0002757797"
[1] " 2 1525.9978611679 45.3663647784 68.4682771320 55800.6149360006 1.0002749247"
[1] " 3 1525.9978611699 45.3663647734 68.4682770954 55800.6626321327 1.0002740696"
[1] " 4 1525.9978611658 45.3663647798 68.4682771807 55800.7103325295 1.0002732143"
[1] " 5 1525.9978611681 45.3663647760 68.4682771288 55800.7580488383 1.0002723589"
[1] " 6 1525.9978611788 45.3663647769 68.4682768710 55800.8057672442 1.0002715038"
[1] " 7 1525.9978611697 45.3663647797 68.4682770847 55800.8534679000 1.0002706483"
[1] " 8 1525.9978611703 45.3663647790 68.4682770717 55800.9011965519 1.0002697926"
[1] " 9 1525.9978611726 45.3663647734 68.4682770288 55800.9489323133 1.0002689368"
[1] " 10 1525.9978611725 45.3663647772 68.4682770196 55800.9966705828 1.0002680810"lam_rank1 = out_rank1$ql$mean %*% t(out_rank1$qf$mean)
ll_train_rank1 = sum(dpois(sim$X, lam_rank1, log = T))
ll_val_rank1 = sum(dpois(sim$Y, lam_rank1, log = T))
rmse_rank1 = sqrt(mean((lam_rank1 - sim$Lam)^2))
data.frame(ll_train = ll_train_rank1, ll_val = ll_val_rank1, rmse = rmse_rank1) ll_train ll_val rmse
1 -37674.75 -37791.54 0.1434967rank-k:
out = ebpmf::ebpmf_point_gamma(X = sim$X, K = K, maxiter.out = 100, verbose = F, fix_g = F)
lam_out = out$qg$qls_mean %*% t(out$qg$qfs_mean)
ll_train_out = sum(dpois(sim$X, lam_out, log = T))
ll_val_out = sum(dpois(sim$Y, lam_out, log = T))
rmse_out = sqrt(mean((lam_out - sim$Lam)^2))
d = length(out$ELBO)
print(sprintf("ELBO monotonically increasing? %s", all(out$ELBO[1:(d-1)] < out$ELBO[2:d])))[1] "ELBO monotonically increasing? TRUE"plot(out$ELBO)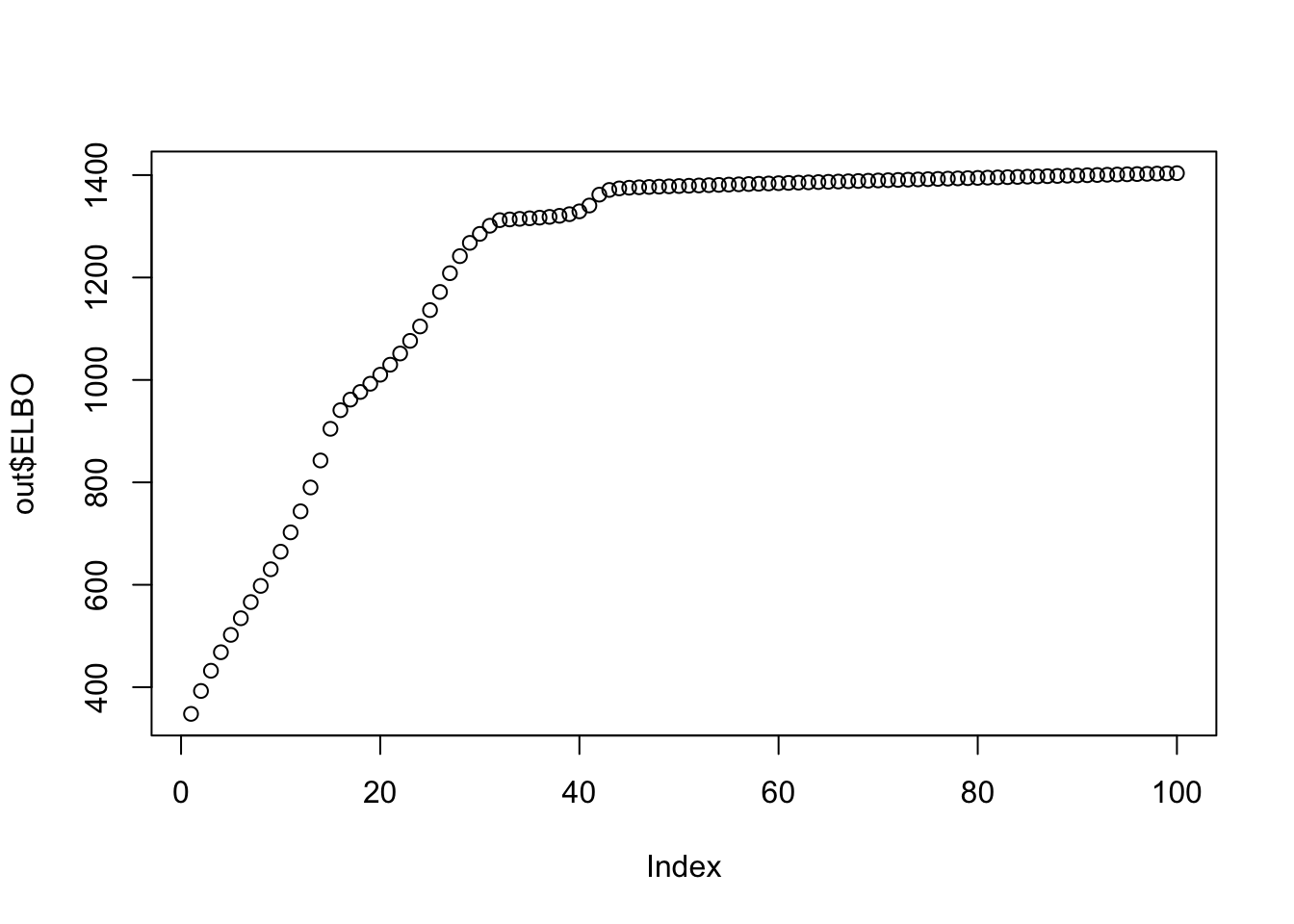
data.frame(ll_train = ll_train_out, ll_val = ll_val_out, rmse = rmse_out) ll_train ll_val rmse
1 -37714.18 -37832.52 0.1671491mle
mle_rankK = NNLM::nnmf(A = sim$X, init = list(W0 = out$qg$qls_mean, H0 = t(out$qg$qfs_mean)),k = K, method = "lee", loss = "mkl", rel.tol = 1e-10, max.iter = 100)
lam_mle_rankK = mle_rankK$W %*% mle_rankK$H
ll_train_mle_rankK = sum(dpois(sim$X, lam_mle_rankK, log = T))
ll_val_mle_rankK = sum(dpois(sim$Y, lam_mle_rankK, log = T))
rmse_mle_rankK = sqrt(mean((lam_mle_rankK - sim$Lam)^2))
data.frame(ll_train = ll_train_mle_rankK, ll_val = ll_val_mle_rankK, rmse = rmse_mle_rankK) ll_train ll_val rmse
1 -36870.87 -38462.89 0.4485559
sessionInfo()R version 3.5.1 (2018-07-02)
Platform: x86_64-apple-darwin15.6.0 (64-bit)
Running under: macOS 10.14
Matrix products: default
BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
locale:
[1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] NNLM_0.4.2 ebpmf_0.1.0 testthat_2.2.1
loaded via a namespace (and not attached):
[1] Rcpp_1.0.2 compiler_3.5.1 git2r_0.25.2
[4] workflowr_1.4.0 prettyunits_1.0.2 remotes_2.1.0
[7] tools_3.5.1 digest_0.6.21 pkgbuild_1.0.3
[10] pkgload_1.0.2 evaluate_0.14 memoise_1.1.0
[13] rlang_0.4.0 cli_1.1.0 rstudioapi_0.10
[16] yaml_2.2.0 xfun_0.8 withr_2.1.2
[19] stringr_1.4.0 knitr_1.25 gtools_3.8.1
[22] desc_1.2.0 fs_1.3.1 devtools_2.2.1.9000
[25] rprojroot_1.3-2 glue_1.3.1 R6_2.4.0
[28] processx_3.3.1 rmarkdown_1.13 sessioninfo_1.1.1
[31] mixsqp_0.1-121 callr_3.2.0 magrittr_1.5
[34] whisker_0.3-2 backports_1.1.5 ps_1.3.0
[37] ellipsis_0.3.0 htmltools_0.3.6 usethis_1.5.1
[40] assertthat_0.2.1 stringi_1.4.3 ebpm_0.0.0.9001
[43] crayon_1.3.4